New Design
Ensembl began in 2000 as a browser for the human genome, but has expanded to host the genomes and associated information for more than 40 species, and the number continues to grow steadily. In 2008, we are pleased to announce our new design, which will greatly increase speed and usability of the website. During the next few months, we ask you to bear with us as the new design will be incorporating more pages to create a comprehensive site. In the meantime, our archive sites will be available as usual for you to compare data with the new release.
From release 51 onwards, Ensembl will run on improved web code with a new visual design:
What We've Changed, and Why
Extensive changes to the web code have been implemented to improve speed and maintainability. In addition, the user interface has been changed substantially.
Design changes
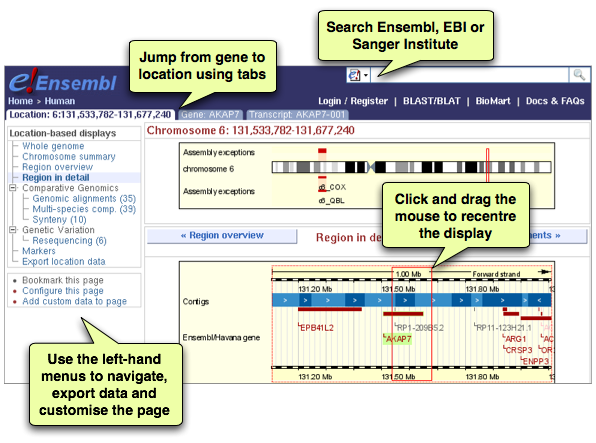
One of the continuing challenges with a site that contains so much data is making it discoverable. In testing sessions we found that users were overlooking the left-hand menu with its links to additional data, perhaps because there was so much on the page already that they didn't expect to find even more!
The main change we have made is to improve the way that users move between pages. The displays have been grouped into Location, Gene, Transcript and Variation and tabs at the top of the page allow the user to switch easily between these sets of views. If you browse to a location on a genome, initially only the Location tab is present, but the others appear as you select specific genes and transcripts to view.
In addition, this tab/menu style of interface has been extended to other pages, including image configuration, help and user accounts, making the site more consistent in appearance and hopefully easier to use.
We addressed the left-hand menu problem in several ways:
- By making the individual displays hold less information, thus hopefully encouraging users to look around for more
- By reducing the visual contrast between the page and its left-hand menu, making it easier for the eye to move away from the central image and onto the menu
- By eliminating non-essential links from the left-hand menu to reduce visual clutter
- By providing a more intuitive list of links that in many cases reveal how much more data is available, e.g. 'Synteny (7 species)'
You will also have noticed that we have changed the colour scheme somewhat, darkening the blue background and extending it to the whole page header, and using the yellow background to draw attention to important parts of the page such as genomic images and information boxes.
Control Panel
The Ensembl website had grown somewhat haphazardly over the last seven years, with features added on an ad hoc basis when required. As a result there were often different interfaces or options available on different pages and configuration menus were often overlooked by users.
We have therefore introduced the Control Panel, a popup window which presents a common interface on all pages and gives access to user-controlled options, including:
- Adding and removing tracks
- Configuring graphical displays, e.g. image size, markup options on sequence displays
- Logging in and managing your user account
- Uploading and managing your own data, including external DAS sources
Other changes
A few things have been left out simply because they are no longer needed. For example, we no longer include families in the search index, as the IDs change from release to release and are therefore not a reliable way to look up genes.
We have also dropped the site maps which linked to each species' genomic views, as the new navigation allows quick and easy access to the same information. A full table of contents is still available for all our online documentation.
To Come...
Unfortunately we haven't been able to port all our old views to the new code yet, so there may be some functionality missing. Please accept our apologies for their absence - we will get around to them in a release or two, and hopefully they will be better than ever. Temporarily missing views include multicontigview and unmapped features in featureview.
Technical details
Performance improvements
These have been achieved in three main ways:
- We have replaced our shared filesystem with shared memory using memcached. This allows us to cache a lot more content and serve it back to the user straight from memory instead of having to read from a disk or access a database.
- Where possible, larger page elements such as genomic images and tables of data are loaded via AJAX. This means that you get most of the page straight away - any section that needs to fetch large amounts of data will display a spinner graphic whilst you wait.
- Changes to the Apache configuration mean that browsers need to make fewer requests when checking for updated content.
- JavaScript and CSS files have been consolidated and compressed so that your browser makes fewer and smaller requests from our server.
- By breaking the display pages into smaller units (see "Design changes", above), it is now possible to return data more quickly - even very large genes such as Titin can be displayed without timing out.
Maintenance issues
Finally, for the benefit of ourselves as well as all the projects who use our software, we have given our drawing and image configuration code a major overhaul so that configuring graphical displays is much easier. The drawing code in particular had hardly been changed since it was first written seven years ago, so was long overdue for updating.
Now, instead of hundreds of glyphset modules and hard-coded configurations, information about track display is stored in the core database and automatically incorporated into the webcode without the need to edit half-a-dozen different files. Also, the drawing code is now pluggable like the main web code, making it even easier to configure your Ensembl-powered site to work just the way you want it.
We have also made species home pages dynamically-generated, instead of static HTML. This is mainly for the benefit of upcoming Ensembl-powered sites which will have a great many species, e.g. bacteria, but will also mean less HTML maintenance for all Ensembl sites.
Inevitably development on this scale has taken a long time, but we hope it has been worth the wait!
Regards,
The Ensembl Team

